Calibration run
Prepare a suitable config.in file; look in
/afs/f9.ijs.si/data/lab/studen/data/20150305/si
for a sample.
pedestalDown
Execute
vmedaq pedestalDown.xml
Prepare a pedestalConfig.in file; look in
/afs/f9.ijs.si/data/lab/studen/data/20150305/si
for a sample. Make sure all paths in pedestalConfig.in are correct
Run observer:
/bin/execute /path/to/pedestal/Config/pedestalConfig.in
pedestalUp
The same; pedestalConfig.in is called pedestalConfig1.in.
extractPedestal
This is in file scripts_processTwoChannelDataToRatio.C which requires
processTwoChannelDataToRatio.config file. There is a copy in
/afs/f9.ijs.si/data/lab/studen/data/20150305/si
and pedestalUp and pedestalDown should point to existing files.
Load the root script;
dataCont dc;
make_pedestal_file(dc, file_down, file_up);
where file_down and file_up is something sensible.
Update config file with pedestal file
In config.in, there are tags for pedestal file where file_down and file_up can
be inserted.
Process real calibration run
The config file for observer run is dataConfig.in. Updating it should be clear.
ROOT part
The config file is processCalib.config and processCalib.C+ should be loaded. Then:
root[] dataQACalib dq
root[] dq.init("/path/to/processCalib.config")
root[] dataQA dd;
root[] processCalibVA(dq,&dd);
Some nice plots are
dd.hSignalVA[0] \\down
dd.hSignalVA[1] \\up
dd.hNoiseVA[0] \\variation of the injection, down
dd.hNoiseVA[1] \\variation of the injection,up
dd.hBaseNoiseVA[0] \\variation of the baseline, down
dd.hBaseNoiseVA[1] \\variation of the baseline, up
Am run analysis
Assume use of pedestals from previous run; if not, repeat steps above to obtain new valid pedestals.
Am observer part
Configuration in a dataConfig.in; check
/afs/f9.ijs.si/data/lab/studen/data/20150309/si/dataConfig.in
I reused config.in from 20150305 calibration run! In 20150305 are also the
then recorded calibration files.
One should make the out directory;
mkdir /path/to/data/out;
eg.
mkdir /afs/f9.ijs.si/data/lab/studen/data/20150309/si/out
Default is not to use pedestals; that is specified in config.in, doPedestal=0.
Am analysis part (for non-pedestal processed files)
Use dataCont infrastructure to match pedestals from 20150305 run and processed
data from 20150309. So, use processTwoChannelDataToRatio.config where pedestal
files (H2 dumps) from 20150305 are used together with just processed 20150309
data.
Then load
root [] .L scripts_processTwoChannelDataToRatio.C+
root [] dataCont dc
root [] dc.init("/path/to/processTwoChannelDataToRatio.config")
root [] set_pedestal(dc,1)
root [] TH2D hdw,hup;
root [] processSettings ps
root [] processAm(dc,hdw,hup,ps)
and hdw (down) and hup (up) hold processed and pedestal subtracted data.
Am alginment
For data alignment, use processGainAm (loaded by default). We'll need:
root[] processData pd;
Insert our processed data;
root[] hdw.Copy(pd.aH)
and calculate gain
root[] processGainAm(pd,ps);
(in ps, you can adjust ranges of fits, fitting parameters, etc.)
Extracting aligned Am histogram
To use alignments; rerun processAm from scripts_processTwoChannelDataToRatio.C
root [] ps.maxE=120;
root [] ps.nBinsE=240;
root [] processAm(dc,hdw,hup,ps,&pd,1) //for up
root [] processAm(dc,hdw,hup,ps,&pd,0) //for down
For 1D spectra; do
root [] hdw.ProjectionY("py",2,512)->Draw();
Am analysis with on-the-fly pedestals
One option is to let the routine calculate pedestal from adjacent channels.
In 20150309/si there is a dataConfig_wPedestal.in which points to config.si in
20150309/si which is tuned for pedestal calculation. Note that eADCPed output
field was added.
To add a default dataConfig_wPedestal_runX.in do
~/software/build/observer_tools/parse_si/scripts/generate_dataConfig.sh\
X /afs/f9.ijs.si/data/lab/studen/data/20150320/si
where X is the target run. Then process the data
~/software/build/observer/bin/execute\
/afs/f9.ijs.si/data/lab/studen/data/20150320/si/dataConfig_wPedestal_runX.in
Am analysis (for pedestal processed files)
In ROOT:
root[] processSettings ps;
root[] processData pd;
Adjust treeMode. For data processed to indivudal ADCs,
treeMode=0. If combined setting is used, ie. if configCombined.in
is used in place of config.in in dataConfig_wPedestal.in, then the
variable treeMode should be set to 1.
root[] ps.treeMode=1;
The processing is done via a single shot:
root[] processRun_tb(
"/afs/f9.ijs.si/data/lab/studen/data/20150320/si/out/Am241_hamamatsuRigidFlex_20150320_g1_s0_s0.out"
,pd,ps)
The final aligned histogram is obtained by:
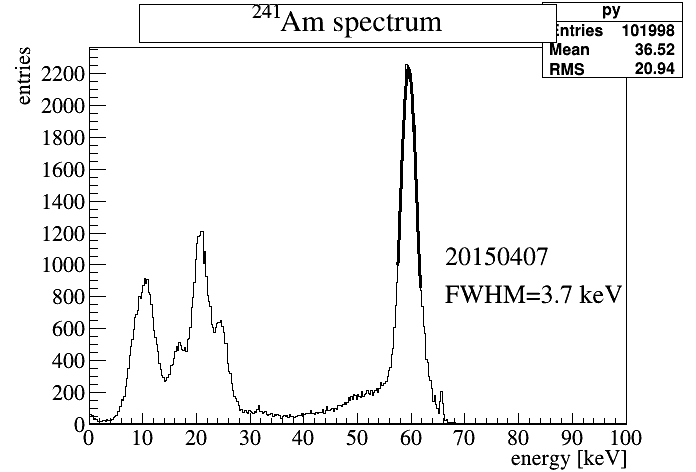
root[] pd.hS.ProjectionY("py",1,128)->Draw();
and standard tools (like Fit Panel) can be used to extract resolution.